
TMIC and the Metabolomics Society
Issue 44 - April 2015
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Apr2015/MetaboNews_Apr2015.htm
 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 44 - April 2015 |
|
CONTENTS: |
|
|
 |
|
 |
|
 |
| Metanomics Health GmbH |
Chenomx
Inc. |
mzCloud
Mass Spectral Database |
 |
Metabolomics Society News |

| |
Metabolomics Spotlight
|
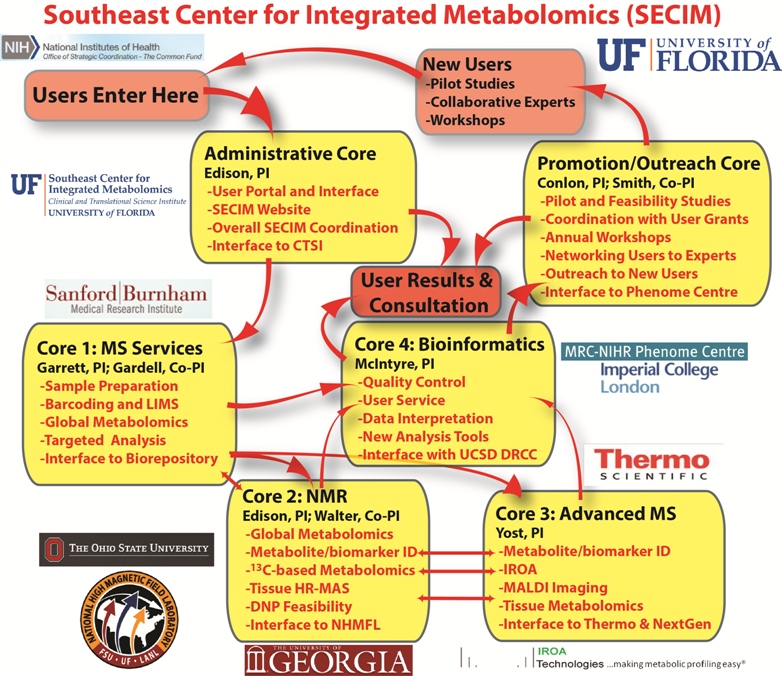
SECIM also has a Promotion and Outreach Core, which has made
a number of instructional videos, offers annual metabolomics
workshops, and provides annual pilot and feasibility grants.
SECIM is currently reviewing applications for the 2015 Pilot
and Feasibility program, and will announce awards soon. The
next SECIM
Metabolomics Workshop is May 11-14, 2015.
For more details and to subscribe to our mailing list, please
visit http://secim.ufl.edu.
| |
MetaboInterview
|
Scientific Coordinator, Metabolomics Platform of the Spanish Biomedical Research Centre in Diabetes and Associated Metabolic Disorders (CIBERDEM) and Assistant Professor at Rovira i Virgili University (Tarragona, Spain) |
|
| Biography Oscar Yanes received his B.A. and Ph.D.
degrees in biochemistry from the Autonomous University
of Barcelona (Spain). In 2007, he joined The Scripps
Center for Metabolomics and Mass Spectrometry (La Jolla,
California) headed by Dr. Gary Siuzdak. Since January
2011, he has served as the scientific coordinator of the
Metabolomics Platform of the Spanish Biomedical Research
Centre in Diabetes and Associated Metabolic Disorders
(CIBERDEM) and Assistant Professor at Rovira i Virgili
University (Tarragona, Spain), where he also leads the
Yanes Lab group (www.yaneslab.com).
He has extensive experience in developing new technologies, methods, and applications in mass spectrometry-based metabolomics. His lab now focuses on understanding metabolic dysregulations in disease by integrating MS and NMR-based metabolomics with other omic platforms. |




 |
Metabolomics Current Contents
|
This section of MetaboNews is
supported by: |
Metabolomics Events
|
| 11-14 May 2015 |
2015 SECIM Workshop The 2nd Annual SECIM Metabolomics Workshop will be
held Monday, May 11 through Thursday, May 14, 2015,
in Gainesville, Florida. Click
here for the event flyer For more information, visit http://secim.ufl.edu/workshops/. |
| 21-22 May 2015 |
2nd Metabolomics - Advances &
Applications in Human Disease Conference Panel: |
| 14-18 Jun 2015 |
3rd Annual Workshop on
Metabolomics The themes in this third year of the workshop are:
|
| 15-16 Jun 2015 |
Informatics and Statistics for
Metabolomics (2015) A poster announcing this workshop can be found here.The workshop will cover many topics ranging from understanding metabolomics technologies, data collection and analysis, using pathway databases, performing pathway analysis, conducting univariate and multivariate statistics, working with metabolomic databases and exploring chemical databases. Participants will be given various data sets and short assignments to assist with the learning process. Target Audience This course is intended for graduate students, post-doctoral fellows, clinical fellows and investigators who are interested in learning about both bioinformatic and cheminformatic tools to analyze and interpret metabolomics data. Prerequisite: Familiarity with R is required. Familiarity can be gained through online activities. You should be familiar with these R concepts (chapters 1-5) or review the past Statistics tutorials provided by CBW. Apply Now Award Opportunities For further details, visit http://bioinformatics.ca/workshops/2015/informatics-and-statistics-metabolomics-2015. |
| 15-18 Jun 2015 |
Metabolomics Summer Workshop This workshop is intended for investigators seeking a solid foundation to expand their research using metabolomics. Sessions include:
For further details, visit http://mrc2.umich.edu/Events_Summer_Workshop.php. |
| 29 Jun to 2 Jul 2015 |
Metabolomics 2015: 11th Annual
Conference of the International Metabolomics
Society You are invited to join us for Metabolomics
2015, the official annual meeting of the
Metabolomics Society. For further details, visit http://metabolomics2015.org. |
| 7-9 Dec 2015 |
MetaboMeeting 2015 Agenda Topics
|
Metabolomics Jobs |
This is a resource for advertising positions in
metabolomics. If you have a job you would like posted in
this newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4 issues (8
weeks) unless the position is filled prior to that date.
Jobs Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Mass Spectrometry Metabolomics Research
Fellow |
University of Birmingham | Birmingham, UK |
27-Mar-2015 |
April 8, 2015 |
Metabolomics
Society Jobs |
| Experimental Officer in NMR Metabolite
Analysis |
University of Birmingham | Birmingham, UK |
27-Mar-2015 |
April 10, 2015 |
Metabolomics
Society Jobs |
| Research Assistant I, Metabolomics Core
Facility |
Sanford-Burnham
Medical Research Institute |
Orlando, Florida, USA | 3-Mar-2015 |
Open until filled |
Metabolomics
Society Jobs |
| Postdoctoral Research Fellow, Fernandez
Laboratory, School of Chemistry and Biochemistry |
Georgia Institute of Technology |
Atlanta, Georgia, USA | 19-Feb-2015 |
Until position is filled |
Georgia Institute of Technology |
| Computational Metabolomics Professor |
Pennsylvania State University |
State College, Pennsylvania, USA |
26-Jan-2015 |
Metabolomics Society Jobs |
|
| Postdoc – Research Animal Scientist |
USDA, Agriculture Research Service, U.S. Meat Animal Research Center |
Clay Center, NE USA | 6-Nov-2014 |
Upon identification of suitable candidate |
Metabolomics Society Jobs |
| 1 year Master project at the Liggins
Institute |
University of Auckland, New Zealand | Auckland, New
Zealand |
20-Oct-2014 |
Metabolomics Society Jobs | |
| Applications Support Scientist (f/m)
Metabolomics-Lipidomics |
Thermo Fisher
Scientific |
EU - European |
16-Jul-2014 |
Thermo
Fisher Scientific |
|
Ian J. Forsythe, M.Sc.
MetaboNewsEditor Department of
Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is published in
partnership between The Metabolomics Innovation
Centre (TMIC, http://www.metabolomicscentre.ca/) and the Metabolomics
Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide metabolomics
community.
|